class: center, middle, title-slide # Tidymodels ## An Overview ### Emil Hvitfeldt ### 2021-06-03 --- <div style = "position:fixed; visibility: hidden"> `$$\require{color}\definecolor{orange}{rgb}{1, 0.603921568627451, 0.301960784313725}$$` `$$\require{color}\definecolor{blue}{rgb}{0.301960784313725, 0.580392156862745, 1}$$` `$$\require{color}\definecolor{pink}{rgb}{0.976470588235294, 0.301960784313725, 1}$$` </div> <script type="text/x-mathjax-config"> MathJax.Hub.Config({ TeX: { Macros: { orange: ["{\\color{orange}{#1}}", 1], blue: ["{\\color{blue}{#1}}", 1], pink: ["{\\color{pink}{#1}}", 1] }, loader: {load: ['[tex]/color']}, tex: {packages: {'[+]': ['color']}} } }); </script> <style> .orange {color: #FF9A4D;} .blue {color: #4D94FF;} .pink {color: #F94DFF;} </style> # About Me - Data Analyst at Teladoc Health - Adjunct Professor at American University teaching statistical machine learning using {tidymodels} - R package developer, about 10 packages on CRAN (textrecipes, themis, paletteer, prismatic, textdata) - Co-author of "Supervised Machine Learning for Text Analysis in R" with Julia Silge - Located in sunny California - Has 3 cats; Presto, Oreo, and Wiggles --- background-image: url(images/cats.png) background-position: center background-size: contain --- background-image: url(hex/tidymodels.png) background-position: 90% 20% background-size: 250px # What is {tidymodels}? - A collection of many packages - Focused on modeling and machine learning - Using tidymodels principles --- class: center # Core packages rsample parsnip recipes tune workflows yardstick dials broom --- background-image: url(diagrams/model.png) background-position: center background-size: contain --- background-image: url(diagrams/model-evaluate.png) background-position: center background-size: contain --- background-image: url(diagrams/preprocess-model-evaluate.png) background-position: center background-size: contain --- background-image: url(diagrams/split-preprocess-model-evaluate.png) background-position: center background-size: contain --- background-image: url(diagrams/full-game.png) background-position: center background-size: contain --- background-image: url(diagrams/full-game-parsnip.png) background-position: center background-size: contain --- # User-facing problems in modeling in R - Data must be a matrix (except when it needs to be a data.frame) - Must use formula or x/y (or both) - Inconsistent naming of arguments (ntree in randomForest, num.trees in ranger) - na.omit explicitly or silently - May or may not accept factors --- # Syntax for Computing Predicted Class Probabilities |Function |Package |Code | |:------------|:------------|:------------------------------------------| |`lda` |`MASS` |`predict(obj)` | |`glm` |`stats` |`predict(obj, type = "response")` | |`gbm` |`gbm` |`predict(obj, type = "response", n.trees)` | |`mda` |`mda` |`predict(obj, type = "posterior")` | |`rpart` |`rpart` |`predict(obj, type = "prob")` | |`Weka` |`RWeka` |`predict(obj, type = "probability")` | |`logitboost` |`LogitBoost` |`predict(obj, type = "raw", nIter)` | --- ## The goals of `parsnip` is... - Decouple the .blue[model classification] from the .orange[computational engine] - Separate the definition of a model from its evaluation - Harmonize argument names - Make consistent predictions (always tibbles with `na.omit = FALSE`) --- # Parsnip usage ```r linear_spec <- lm(mpg ~ disp + drat + qsec, data = mtcars) ``` --- # Parsnip usage .pull-left[ ```r library(parsnip) linear_spec <- linear_reg() %>% set_mode("regression") %>% set_engine("lm") linear_spec ``` ``` ## Linear Regression Model Specification (regression) ## ## Computational engine: lm ``` ] .pull-right[ ```r fit_lm <- linear_spec %>% fit(mpg ~ disp + drat + qsec, data = mtcars) fit_lm ``` ``` ## parsnip model object ## ## Fit time: 1ms ## ## Call: ## stats::lm(formula = mpg ~ disp + drat + qsec, data = data) ## ## Coefficients: ## (Intercept) disp drat qsec ## 11.52439 -0.03136 2.39184 0.40340 ``` ] --- # Tidy prediction .pull-left[ Consistent Predictions ] .pull-right[ ```r predict(fit_lm, mtcars) ``` ``` ## # A tibble: 32 x 1 ## .pred ## <dbl> ## 1 22.5 ## 2 22.7 ## 3 24.9 ## 4 18.6 ## 5 14.6 ## 6 19.2 ## 7 14.3 ## 8 23.8 ## 9 25.7 ## 10 23.0 ## # … with 22 more rows ``` ] --- # Parsnip models .center[  ] --- .center[ 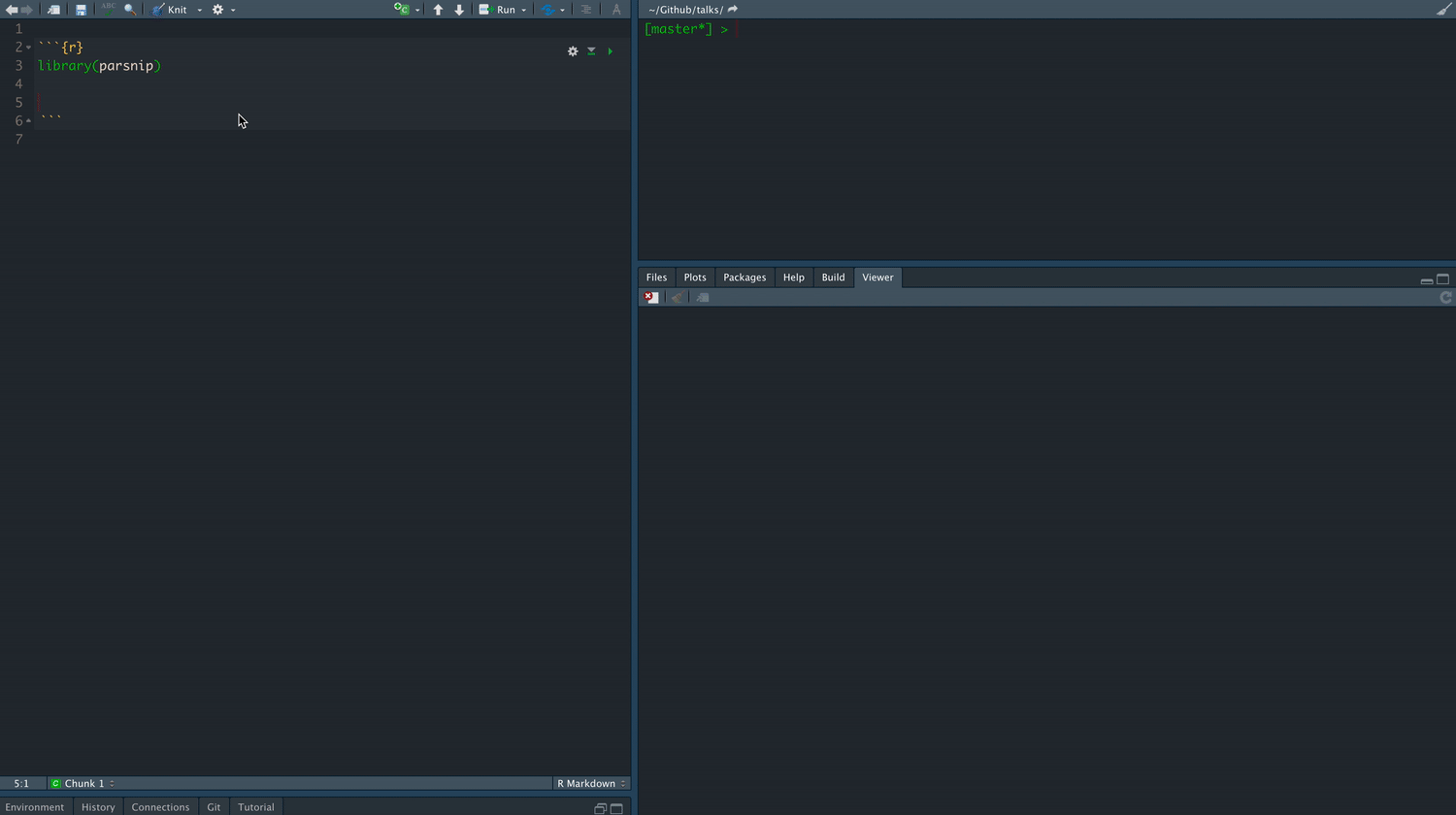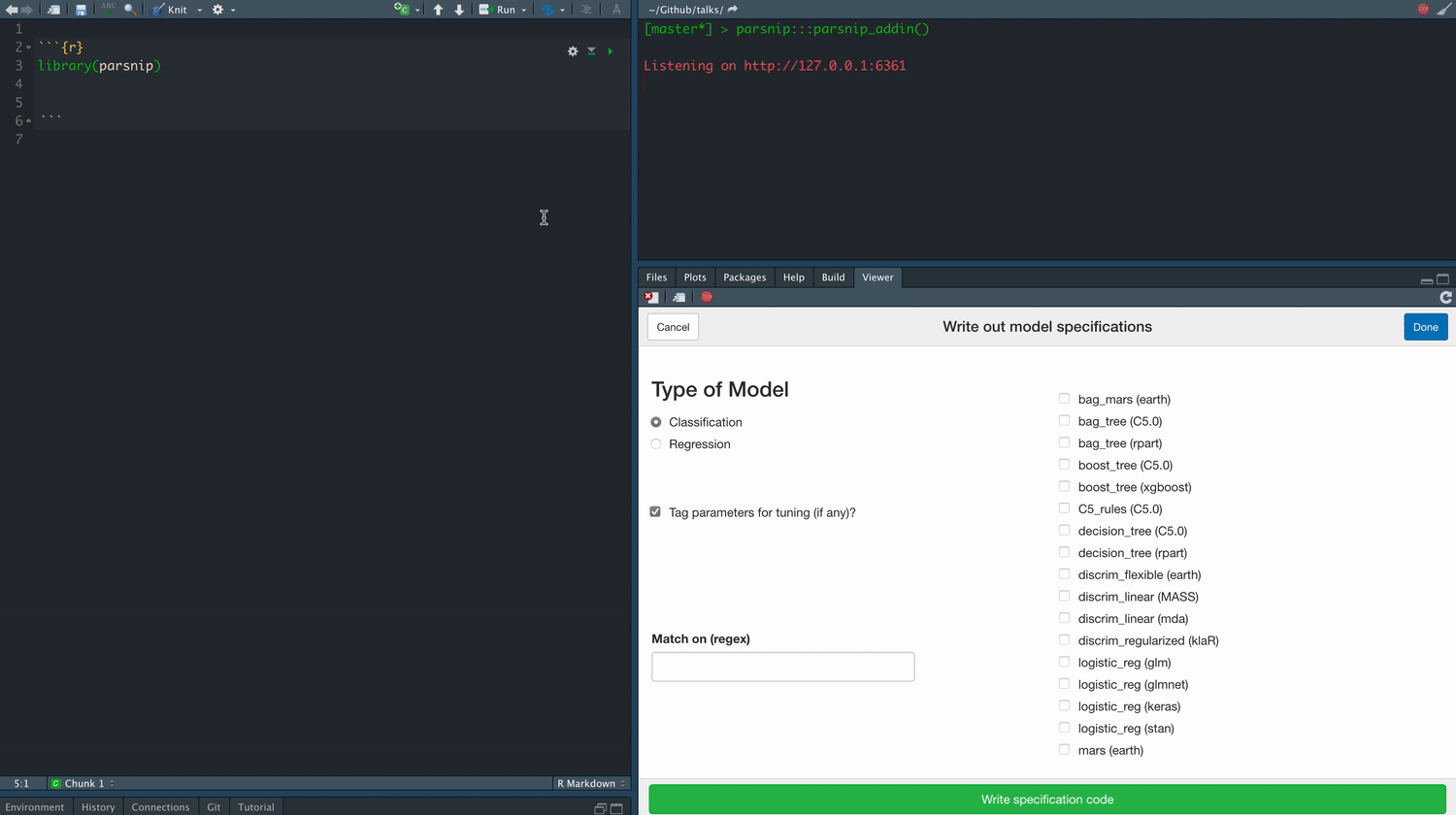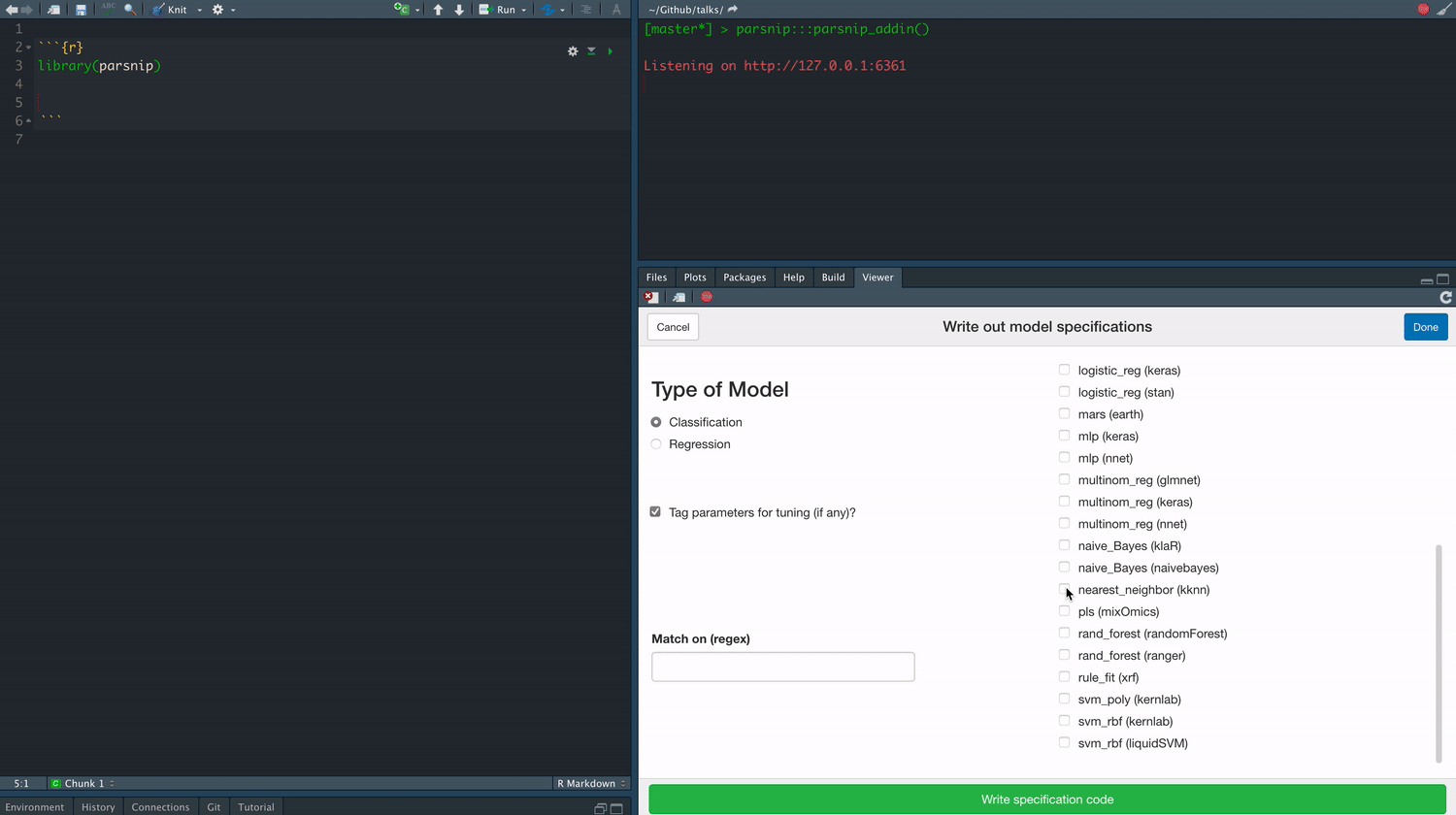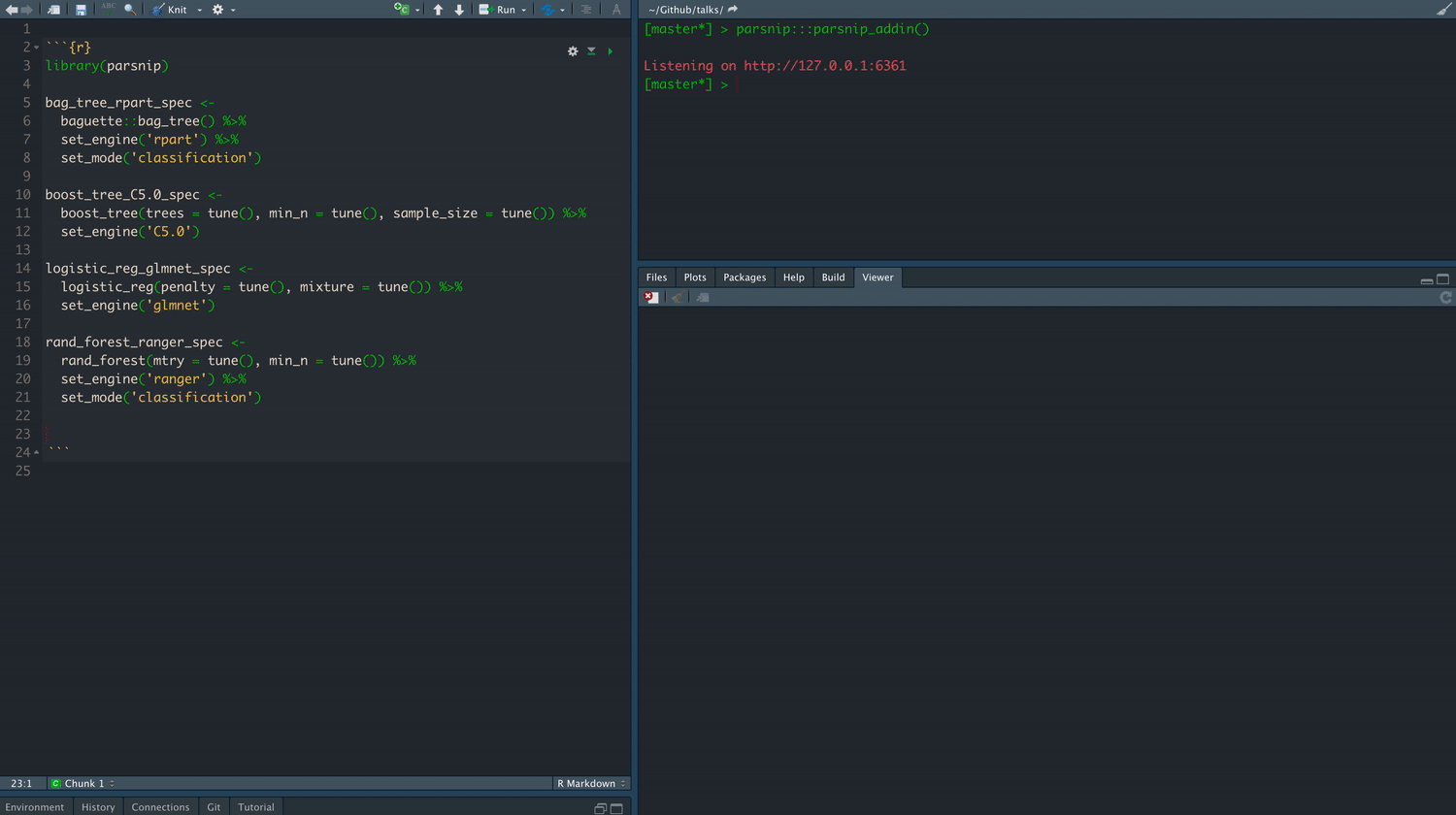 ] --- background-image: url(diagrams/full-game-broom.png) background-position: center background-size: contain --- # broom broom summarizes key information about models in tidy `tibble()`s. broom provides three verbs to make it convenient to interact with model objects: - `tidy()` summarizes information about model components - `glance()` reports information about the entire model - `augment()` adds information about observations to a data set --- # lm fit object ```r fit_lm$fit ``` ``` ## ## Call: ## stats::lm(formula = mpg ~ disp + drat + qsec, data = data) ## ## Coefficients: ## (Intercept) disp drat qsec ## 11.52439 -0.03136 2.39184 0.40340 ``` --- # lm fit object ```r summary(fit_lm$fit) ``` ``` ## ## Call: ## stats::lm(formula = mpg ~ disp + drat + qsec, data = data) ## ## Residuals: ## Min 1Q Median 3Q Max ## -5.4681 -2.0867 -0.7474 1.1838 6.4843 ## ## Coefficients: ## Estimate Std. Error t value Pr(>|t|) ## (Intercept) 11.524390 11.887430 0.969 0.340616 ## disp -0.031364 0.007809 -4.017 0.000402 *** ## drat 2.391842 1.637812 1.460 0.155314 ## qsec 0.403403 0.382875 1.054 0.301067 ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ## ## Residual standard error: 3.226 on 28 degrees of freedom ## Multiple R-squared: 0.7413, Adjusted R-squared: 0.7135 ## F-statistic: 26.74 on 3 and 28 DF, p-value: 2.274e-08 ``` --- # lm fit object ```r coefficients(fit_lm$fit) ``` ``` ## (Intercept) disp drat qsec ## 11.52439035 -0.03136425 2.39184212 0.40340322 ``` --- # lm fit object ```r tidy(fit_lm) ``` ``` ## # A tibble: 4 x 5 ## term estimate std.error statistic p.value ## <chr> <dbl> <dbl> <dbl> <dbl> ## 1 (Intercept) 11.5 11.9 0.969 0.341 ## 2 disp -0.0314 0.00781 -4.02 0.000402 ## 3 drat 2.39 1.64 1.46 0.155 ## 4 qsec 0.403 0.383 1.05 0.301 ``` --- # lm fit object ```r glance(fit_lm) ``` ``` ## # A tibble: 1 x 12 ## r.squared adj.r.squared sigma statistic p.value df logLik AIC BIC ## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> ## 1 0.741 0.714 3.23 26.7 0.0000000227 3 -80.7 171. 179. ## # … with 3 more variables: deviance <dbl>, df.residual <int>, nobs <int> ``` --- # lm fit object ```r augment(fit_lm, new_data = mtcars[1:5, ]) ``` ``` ## # A tibble: 5 x 13 ## mpg cyl disp hp drat wt qsec vs am gear carb .pred .resid ## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> ## 1 21 6 160 110 3.9 2.62 16.5 0 1 4 4 22.5 -1.47 ## 2 21 6 160 110 3.9 2.88 17.0 0 1 4 4 22.7 -1.70 ## 3 22.8 4 108 93 3.85 2.32 18.6 1 1 4 1 24.9 -2.05 ## 4 21.4 6 258 110 3.08 3.22 19.4 1 0 3 1 18.6 2.76 ## 5 18.7 8 360 175 3.15 3.44 17.0 0 0 3 2 14.6 4.07 ``` --- background-image: url(diagrams/full-game-yardstick.png) background-position: center background-size: contain --- # yardstick You now have a model that can produce prediction How do you measure the performance? --- # Yardstick Classification data ```r data(two_class_example, package = "modeldata") two_class_example ``` ``` ## truth Class1 Class2 predicted ## 1 Class2 3.589243e-03 9.964108e-01 Class2 ## 2 Class1 6.786211e-01 3.213789e-01 Class1 ## 3 Class2 1.108935e-01 8.891065e-01 Class2 ## 4 Class1 7.351617e-01 2.648383e-01 Class1 ## 5 Class2 1.623996e-02 9.837600e-01 Class2 ## 6 Class1 9.992751e-01 7.249286e-04 Class1 ## 7 Class1 9.992011e-01 7.988510e-04 Class1 ## 8 Class1 8.123520e-01 1.876480e-01 Class1 ## 9 Class2 4.570373e-01 5.429627e-01 Class2 ## 10 Class2 9.763773e-02 9.023623e-01 Class2 ## 11 Class2 9.046455e-01 9.535452e-02 Class1 ## 12 Class1 2.979792e-01 7.020208e-01 Class2 ## 13 Class1 9.945820e-01 5.418045e-03 Class1 ## 14 Class2 8.511498e-01 1.488502e-01 Class1 ## 15 Class1 9.999965e-01 3.492550e-06 Class1 ## 16 Class1 9.919163e-01 8.083723e-03 Class1 ## 17 Class2 4.250834e-04 9.995749e-01 Class2 ## 18 Class2 1.149549e-01 8.850451e-01 Class2 ## 19 Class1 8.259703e-01 1.740297e-01 Class1 ## 20 Class2 6.836781e-02 9.316322e-01 Class2 ## 21 Class1 9.350963e-01 6.490371e-02 Class1 ## 22 Class1 9.999720e-01 2.798780e-05 Class1 ## 23 Class2 2.220960e-01 7.779040e-01 Class2 ## 24 Class2 2.076886e-01 7.923114e-01 Class2 ## 25 Class2 2.395558e-02 9.760444e-01 Class2 ## 26 Class1 9.998816e-01 1.184037e-04 Class1 ## 27 Class1 9.811623e-01 1.883772e-02 Class1 ## 28 Class1 9.835188e-01 1.648122e-02 Class1 ## 29 Class2 5.225484e-03 9.947745e-01 Class2 ## 30 Class2 4.214534e-02 9.578547e-01 Class2 ## 31 Class2 2.905598e-03 9.970944e-01 Class2 ## 32 Class1 9.978972e-01 2.102816e-03 Class1 ## 33 Class2 8.299198e-01 1.700802e-01 Class1 ## 34 Class1 8.055859e-01 1.944141e-01 Class1 ## 35 Class2 3.295137e-05 9.999670e-01 Class2 ## 36 Class2 1.612067e-04 9.998388e-01 Class2 ## 37 Class2 1.199105e-01 8.800895e-01 Class2 ## 38 Class1 8.222818e-02 9.177718e-01 Class2 ## 39 Class1 9.234837e-01 7.651630e-02 Class1 ## 40 Class2 9.966767e-03 9.900332e-01 Class2 ## 41 Class1 9.439622e-01 5.603781e-02 Class1 ## 42 Class2 1.592083e-02 9.840792e-01 Class2 ## 43 Class1 9.223776e-03 9.907762e-01 Class2 ## 44 Class2 2.360050e-03 9.976399e-01 Class2 ## 45 Class2 7.474391e-03 9.925256e-01 Class2 ## 46 Class2 6.738986e-01 3.261014e-01 Class1 ## 47 Class2 6.333998e-02 9.366600e-01 Class2 ## 48 Class2 5.023866e-03 9.949761e-01 Class2 ## 49 Class2 1.810806e-02 9.818919e-01 Class2 ## 50 Class1 9.912743e-01 8.725662e-03 Class1 ## 51 Class1 8.945705e-01 1.054295e-01 Class1 ## 52 Class1 9.961046e-01 3.895397e-03 Class1 ## 53 Class1 8.429632e-01 1.570368e-01 Class1 ## 54 Class1 9.973947e-01 2.605314e-03 Class1 ## 55 Class2 5.019505e-01 4.980495e-01 Class1 ## 56 Class1 9.838037e-01 1.619629e-02 Class1 ## 57 Class2 1.221231e-02 9.877877e-01 Class2 ## 58 Class2 1.328124e-03 9.986719e-01 Class2 ## 59 Class2 9.365285e-03 9.906347e-01 Class2 ## 60 Class2 1.296095e-02 9.870390e-01 Class2 ## 61 Class2 3.884927e-02 9.611507e-01 Class2 ## 62 Class1 9.992216e-01 7.783687e-04 Class1 ## 63 Class1 9.623572e-01 3.764278e-02 Class1 ## 64 Class1 9.840572e-01 1.594280e-02 Class1 ## 65 Class1 9.981712e-01 1.828799e-03 Class1 ## 66 Class1 9.986848e-01 1.315244e-03 Class1 ## 67 Class1 9.963867e-01 3.613335e-03 Class1 ## 68 Class2 5.825569e-02 9.417443e-01 Class2 ## 69 Class2 1.173739e-03 9.988263e-01 Class2 ## 70 Class2 7.943867e-01 2.056133e-01 Class1 ## 71 Class1 9.988568e-01 1.143212e-03 Class1 ## 72 Class2 5.815015e-01 4.184985e-01 Class1 ## 73 Class1 9.979431e-01 2.056906e-03 Class1 ## 74 Class2 5.522745e-01 4.477255e-01 Class1 ## 75 Class1 9.847017e-01 1.529833e-02 Class1 ## 76 Class2 1.429375e-05 9.999857e-01 Class2 ## 77 Class1 1.009436e-01 8.990564e-01 Class2 ## 78 Class2 8.561391e-01 1.438609e-01 Class1 ## 79 Class2 4.074602e-01 5.925398e-01 Class2 ## 80 Class1 3.139248e-01 6.860752e-01 Class2 ## 81 Class1 6.019319e-01 3.980681e-01 Class1 ## 82 Class2 3.630955e-02 9.636904e-01 Class2 ## 83 Class2 3.096082e-01 6.903918e-01 Class2 ## 84 Class1 9.482744e-01 5.172563e-02 Class1 ## 85 Class1 9.844007e-01 1.559931e-02 Class1 ## 86 Class1 9.794736e-01 2.052643e-02 Class1 ## 87 Class2 2.375451e-05 9.999762e-01 Class2 ## 88 Class1 7.990447e-01 2.009553e-01 Class1 ## 89 Class2 8.035155e-05 9.999196e-01 Class2 ## 90 Class1 8.534425e-01 1.465575e-01 Class1 ## 91 Class1 9.964761e-01 3.523859e-03 Class1 ## 92 Class2 5.757130e-02 9.424287e-01 Class2 ## 93 Class2 1.033265e-04 9.998967e-01 Class2 ## 94 Class1 1.367068e-01 8.632932e-01 Class2 ## 95 Class1 9.995170e-01 4.829604e-04 Class1 ## 96 Class1 9.863939e-01 1.360610e-02 Class1 ## 97 Class1 9.674599e-01 3.254012e-02 Class1 ## 98 Class1 1.474711e-01 8.525289e-01 Class2 ## 99 Class2 5.895584e-05 9.999410e-01 Class2 ## 100 Class1 9.920976e-01 7.902404e-03 Class1 ## 101 Class2 1.353716e-01 8.646284e-01 Class2 ## 102 Class1 9.773557e-01 2.264430e-02 Class1 ## 103 Class2 2.371418e-02 9.762858e-01 Class2 ## 104 Class1 9.781376e-01 2.186243e-02 Class1 ## 105 Class2 3.977469e-01 6.022531e-01 Class2 ## 106 Class2 1.900919e-01 8.099081e-01 Class2 ## 107 Class2 8.920351e-02 9.107965e-01 Class2 ## 108 Class2 4.141195e-04 9.995859e-01 Class2 ## 109 Class1 9.983074e-01 1.692559e-03 Class1 ## 110 Class1 9.998535e-01 1.465234e-04 Class1 ## 111 Class2 8.463544e-01 1.536456e-01 Class1 ## 112 Class2 9.039022e-01 9.609781e-02 Class1 ## 113 Class2 1.157239e-01 8.842761e-01 Class2 ## 114 Class1 9.840912e-01 1.590885e-02 Class1 ## 115 Class2 1.367840e-03 9.986322e-01 Class2 ## 116 Class1 9.919245e-01 8.075476e-03 Class1 ## 117 Class2 2.294874e-02 9.770513e-01 Class2 ## 118 Class2 2.612947e-01 7.387053e-01 Class2 ## 119 Class2 1.707507e-01 8.292493e-01 Class2 ## 120 Class2 8.376660e-03 9.916233e-01 Class2 ## 121 Class1 9.377396e-01 6.226035e-02 Class1 ## 122 Class2 5.776096e-01 4.223904e-01 Class1 ## 123 Class1 9.333508e-01 6.664924e-02 Class1 ## 124 Class1 6.935601e-01 3.064399e-01 Class1 ## 125 Class1 8.108011e-01 1.891989e-01 Class1 ## 126 Class2 6.861017e-03 9.931390e-01 Class2 ## 127 Class2 1.445695e-01 8.554305e-01 Class2 ## 128 Class1 6.100306e-01 3.899694e-01 Class1 ## 129 Class2 6.564836e-04 9.993435e-01 Class2 ## 130 Class1 8.818895e-01 1.181105e-01 Class1 ## 131 Class2 5.760855e-04 9.994239e-01 Class2 ## 132 Class2 1.121137e-01 8.878863e-01 Class2 ## 133 Class2 2.772588e-03 9.972274e-01 Class2 ## 134 Class1 9.743441e-01 2.565593e-02 Class1 ## 135 Class1 9.830390e-01 1.696097e-02 Class1 ## 136 Class2 5.050952e-01 4.949048e-01 Class1 ## 137 Class1 9.993264e-01 6.735776e-04 Class1 ## 138 Class2 5.902088e-01 4.097912e-01 Class1 ## 139 Class2 1.627430e-01 8.372570e-01 Class2 ## 140 Class1 9.934170e-01 6.582991e-03 Class1 ## 141 Class1 9.998906e-01 1.094164e-04 Class1 ## 142 Class2 7.041413e-04 9.992959e-01 Class2 ## 143 Class1 9.999103e-01 8.974970e-05 Class1 ## 144 Class2 8.005191e-01 1.994809e-01 Class1 ## 145 Class1 9.928700e-01 7.129975e-03 Class1 ## 146 Class1 9.647968e-01 3.520322e-02 Class1 ## 147 Class1 9.978288e-01 2.171183e-03 Class1 ## 148 Class2 5.203140e-01 4.796860e-01 Class1 ## 149 Class1 9.927599e-01 7.240124e-03 Class1 ## 150 Class1 9.985040e-01 1.496046e-03 Class1 ## 151 Class1 9.801404e-01 1.985961e-02 Class1 ## 152 Class1 6.551637e-01 3.448363e-01 Class1 ## 153 Class1 9.461352e-01 5.386485e-02 Class1 ## 154 Class2 5.777406e-01 4.222594e-01 Class1 ## 155 Class2 4.255912e-03 9.957441e-01 Class2 ## 156 Class1 7.266243e-01 2.733757e-01 Class1 ## 157 Class2 1.285994e-04 9.998714e-01 Class2 ## 158 Class2 1.890781e-01 8.109219e-01 Class2 ## 159 Class2 8.020915e-01 1.979085e-01 Class1 ## 160 Class2 4.666482e-04 9.995334e-01 Class2 ## 161 Class2 1.149106e-01 8.850894e-01 Class2 ## 162 Class1 9.955102e-01 4.489843e-03 Class1 ## 163 Class2 3.256008e-02 9.674399e-01 Class2 ## 164 Class1 9.152757e-01 8.472428e-02 Class1 ## 165 Class2 6.483872e-02 9.351613e-01 Class2 ## 166 Class1 1.062401e-01 8.937599e-01 Class2 ## 167 Class1 9.995734e-01 4.265779e-04 Class1 ## 168 Class2 1.403864e-02 9.859614e-01 Class2 ## 169 Class2 8.565222e-02 9.143478e-01 Class2 ## 170 Class2 5.180212e-01 4.819788e-01 Class1 ## 171 Class1 9.623505e-01 3.764947e-02 Class1 ## 172 Class1 8.374863e-01 1.625137e-01 Class1 ## 173 Class2 5.594949e-01 4.405051e-01 Class1 ## 174 Class1 9.798853e-01 2.011465e-02 Class1 ## 175 Class1 9.673368e-01 3.266319e-02 Class1 ## 176 Class2 3.467400e-04 9.996533e-01 Class2 ## 177 Class1 8.713566e-01 1.286434e-01 Class1 ## 178 Class2 8.193828e-01 1.806172e-01 Class1 ## 179 Class2 1.241546e-03 9.987585e-01 Class2 ## 180 Class2 4.798626e-01 5.201374e-01 Class2 ## 181 Class2 9.283173e-01 7.168272e-02 Class1 ## 182 Class2 5.813991e-06 9.999942e-01 Class2 ## 183 Class1 9.993158e-01 6.841886e-04 Class1 ## 184 Class2 3.257151e-01 6.742849e-01 Class2 ## 185 Class2 1.281438e-01 8.718562e-01 Class2 ## 186 Class1 3.344800e-01 6.655200e-01 Class2 ## 187 Class2 1.431378e-01 8.568622e-01 Class2 ## 188 Class1 8.727913e-01 1.272087e-01 Class1 ## 189 Class2 6.966261e-03 9.930337e-01 Class2 ## 190 Class1 9.115189e-01 8.848105e-02 Class1 ## 191 Class1 8.930295e-01 1.069705e-01 Class1 ## 192 Class1 9.982940e-01 1.706036e-03 Class1 ## 193 Class2 1.519918e-03 9.984801e-01 Class2 ## 194 Class1 9.790722e-01 2.092778e-02 Class1 ## 195 Class1 9.659130e-01 3.408703e-02 Class1 ## 196 Class1 8.909204e-01 1.090796e-01 Class1 ## 197 Class2 7.053912e-02 9.294609e-01 Class2 ## 198 Class1 9.925175e-01 7.482477e-03 Class1 ## 199 Class2 2.337041e-01 7.662959e-01 Class2 ## 200 Class1 9.996544e-01 3.455961e-04 Class1 ## 201 Class1 9.373531e-01 6.264694e-02 Class1 ## 202 Class1 3.269821e-01 6.730179e-01 Class2 ## 203 Class1 3.293320e-01 6.706680e-01 Class2 ## 204 Class2 8.699437e-05 9.999130e-01 Class2 ## 205 Class1 9.987389e-01 1.261139e-03 Class1 ## 206 Class2 5.532678e-01 4.467322e-01 Class1 ## 207 Class1 9.705903e-01 2.940971e-02 Class1 ## 208 Class2 6.497331e-02 9.350267e-01 Class2 ## 209 Class1 9.992902e-01 7.098346e-04 Class1 ## 210 Class2 4.244222e-02 9.575578e-01 Class2 ## 211 Class2 1.963028e-03 9.980370e-01 Class2 ## 212 Class1 9.887467e-01 1.125332e-02 Class1 ## 213 Class2 7.845351e-01 2.154649e-01 Class1 ## 214 Class1 9.771202e-01 2.287982e-02 Class1 ## 215 Class2 1.141131e-02 9.885887e-01 Class2 ## 216 Class2 4.818014e-01 5.181986e-01 Class2 ## 217 Class1 9.932526e-01 6.747379e-03 Class1 ## 218 Class1 9.991848e-01 8.152166e-04 Class1 ## 219 Class1 9.985262e-01 1.473830e-03 Class1 ## 220 Class2 2.810221e-03 9.971898e-01 Class2 ## 221 Class1 7.943030e-01 2.056970e-01 Class1 ## 222 Class1 9.886239e-01 1.137606e-02 Class1 ## 223 Class1 9.785505e-01 2.144949e-02 Class1 ## 224 Class2 5.274166e-01 4.725834e-01 Class1 ## 225 Class2 9.364702e-01 6.352982e-02 Class1 ## 226 Class2 1.404483e-01 8.595517e-01 Class2 ## 227 Class2 1.080119e-01 8.919881e-01 Class2 ## 228 Class2 2.237141e-03 9.977629e-01 Class2 ## 229 Class1 9.370266e-01 6.297345e-02 Class1 ## 230 Class2 7.478977e-01 2.521023e-01 Class1 ## 231 Class2 5.833827e-01 4.166173e-01 Class1 ## 232 Class2 7.101063e-03 9.928989e-01 Class2 ## 233 Class1 9.656067e-01 3.439335e-02 Class1 ## 234 Class2 3.437327e-01 6.562673e-01 Class2 ## 235 Class1 9.988154e-01 1.184578e-03 Class1 ## 236 Class1 6.656975e-01 3.343025e-01 Class1 ## 237 Class2 8.782881e-03 9.912171e-01 Class2 ## 238 Class2 2.836697e-01 7.163303e-01 Class2 ## 239 Class2 1.216854e-05 9.999878e-01 Class2 ## 240 Class2 6.745976e-01 3.254024e-01 Class1 ## 241 Class2 9.729242e-01 2.707584e-02 Class1 ## 242 Class2 2.341568e-01 7.658432e-01 Class2 ## 243 Class1 3.511709e-01 6.488291e-01 Class2 ## 244 Class2 6.642630e-01 3.357370e-01 Class1 ## 245 Class2 7.544818e-01 2.455182e-01 Class1 ## 246 Class2 3.832547e-02 9.616745e-01 Class2 ## 247 Class1 9.980666e-01 1.933422e-03 Class1 ## 248 Class1 9.972076e-01 2.792442e-03 Class1 ## 249 Class1 9.517809e-01 4.821908e-02 Class1 ## 250 Class1 9.994148e-01 5.851958e-04 Class1 ## 251 Class1 9.838027e-01 1.619732e-02 Class1 ## 252 Class1 9.871789e-01 1.282109e-02 Class1 ## 253 Class1 4.528194e-01 5.471806e-01 Class2 ## 254 Class1 9.999054e-01 9.462044e-05 Class1 ## 255 Class2 3.054508e-01 6.945492e-01 Class2 ## 256 Class2 9.934345e-01 6.565525e-03 Class1 ## 257 Class2 6.658272e-02 9.334173e-01 Class2 ## 258 Class1 9.961782e-01 3.821759e-03 Class1 ## 259 Class2 5.970668e-05 9.999403e-01 Class2 ## 260 Class2 7.291951e-03 9.927080e-01 Class2 ## 261 Class2 5.922113e-06 9.999941e-01 Class2 ## 262 Class1 7.082959e-01 2.917041e-01 Class1 ## 263 Class2 1.138276e-02 9.886172e-01 Class2 ## 264 Class1 2.218838e-01 7.781162e-01 Class2 ## 265 Class2 9.625496e-05 9.999037e-01 Class2 ## 266 Class1 9.449979e-01 5.500206e-02 Class1 ## 267 Class1 8.972511e-01 1.027489e-01 Class1 ## 268 Class1 8.951949e-01 1.048051e-01 Class1 ## 269 Class2 6.493605e-01 3.506395e-01 Class1 ## 270 Class2 8.924836e-01 1.075164e-01 Class1 ## 271 Class2 2.104593e-01 7.895407e-01 Class2 ## 272 Class2 3.196424e-02 9.680358e-01 Class2 ## 273 Class2 3.803456e-03 9.961965e-01 Class2 ## 274 Class1 8.838073e-01 1.161927e-01 Class1 ## 275 Class1 7.627046e-01 2.372954e-01 Class1 ## 276 Class1 9.199385e-01 8.006150e-02 Class1 ## 277 Class1 6.177250e-01 3.822750e-01 Class1 ## 278 Class1 3.139262e-01 6.860738e-01 Class2 ## 279 Class1 7.867773e-01 2.132227e-01 Class1 ## 280 Class1 9.998949e-01 1.050660e-04 Class1 ## 281 Class1 5.775524e-01 4.224476e-01 Class1 ## 282 Class1 9.973104e-01 2.689642e-03 Class1 ## 283 Class2 2.724634e-01 7.275366e-01 Class2 ## 284 Class2 4.527164e-03 9.954728e-01 Class2 ## 285 Class2 1.051525e-01 8.948475e-01 Class2 ## 286 Class1 9.277335e-01 7.226653e-02 Class1 ## 287 Class1 2.549027e-01 7.450973e-01 Class2 ## 288 Class1 9.810585e-01 1.894149e-02 Class1 ## 289 Class1 9.858894e-01 1.411064e-02 Class1 ## 290 Class1 9.984481e-01 1.551861e-03 Class1 ## 291 Class2 9.923249e-02 9.007675e-01 Class2 ## 292 Class2 1.714900e-03 9.982851e-01 Class2 ## 293 Class1 9.555918e-01 4.440817e-02 Class1 ## 294 Class2 6.594889e-02 9.340511e-01 Class2 ## 295 Class1 9.948170e-01 5.183043e-03 Class1 ## 296 Class1 7.923354e-01 2.076646e-01 Class1 ## 297 Class1 9.834037e-01 1.659627e-02 Class1 ## 298 Class2 1.345976e-04 9.998654e-01 Class2 ## 299 Class2 6.970622e-01 3.029378e-01 Class1 ## 300 Class2 1.688861e-04 9.998311e-01 Class2 ## 301 Class2 1.748054e-01 8.251946e-01 Class2 ## 302 Class2 6.708014e-02 9.329199e-01 Class2 ## 303 Class1 9.996746e-01 3.254426e-04 Class1 ## 304 Class1 9.768781e-01 2.312192e-02 Class1 ## 305 Class2 8.311658e-03 9.916883e-01 Class2 ## 306 Class1 5.808967e-01 4.191033e-01 Class1 ## 307 Class2 1.220275e-01 8.779725e-01 Class2 ## 308 Class2 1.245746e-01 8.754254e-01 Class2 ## 309 Class1 9.894124e-01 1.058759e-02 Class1 ## 310 Class1 9.939475e-01 6.052468e-03 Class1 ## 311 Class2 1.583306e-01 8.416694e-01 Class2 ## 312 Class2 1.704562e-02 9.829544e-01 Class2 ## 313 Class1 9.351591e-01 6.484087e-02 Class1 ## 314 Class2 3.670582e-02 9.632942e-01 Class2 ## 315 Class1 8.776227e-01 1.223773e-01 Class1 ## 316 Class2 4.906213e-01 5.093787e-01 Class2 ## 317 Class2 4.375222e-01 5.624778e-01 Class2 ## 318 Class1 2.564605e-01 7.435395e-01 Class2 ## 319 Class2 8.071665e-03 9.919283e-01 Class2 ## 320 Class1 6.917256e-01 3.082744e-01 Class1 ## 321 Class1 8.287357e-01 1.712643e-01 Class1 ## 322 Class2 5.768219e-05 9.999423e-01 Class2 ## 323 Class1 9.971476e-01 2.852433e-03 Class1 ## 324 Class1 1.225460e-01 8.774540e-01 Class2 ## 325 Class2 9.908362e-01 9.163805e-03 Class1 ## 326 Class2 1.200283e-01 8.799717e-01 Class2 ## 327 Class1 4.807882e-01 5.192118e-01 Class2 ## 328 Class1 9.852943e-01 1.470570e-02 Class1 ## 329 Class1 8.613978e-01 1.386022e-01 Class1 ## 330 Class2 6.680358e-01 3.319642e-01 Class1 ## 331 Class1 9.997196e-01 2.804330e-04 Class1 ## 332 Class2 2.403744e-01 7.596256e-01 Class2 ## 333 Class1 9.993721e-01 6.279376e-04 Class1 ## 334 Class1 9.987801e-01 1.219866e-03 Class1 ## 335 Class2 5.358960e-02 9.464104e-01 Class2 ## 336 Class2 5.350975e-03 9.946490e-01 Class2 ## 337 Class1 3.710924e-01 6.289076e-01 Class2 ## 338 Class2 6.812940e-01 3.187060e-01 Class1 ## 339 Class1 8.230679e-01 1.769321e-01 Class1 ## 340 Class2 4.503065e-06 9.999955e-01 Class2 ## 341 Class2 4.047803e-02 9.595220e-01 Class2 ## 342 Class2 2.224813e-02 9.777519e-01 Class2 ## 343 Class2 7.546886e-04 9.992453e-01 Class2 ## 344 Class1 9.875471e-01 1.245293e-02 Class1 ## 345 Class1 9.995590e-01 4.410121e-04 Class1 ## 346 Class2 2.110135e-01 7.889865e-01 Class2 ## 347 Class1 9.979229e-01 2.077054e-03 Class1 ## 348 Class2 1.861068e-01 8.138932e-01 Class2 ## 349 Class2 7.203792e-01 2.796208e-01 Class1 ## 350 Class1 9.998946e-01 1.054150e-04 Class1 ## 351 Class1 9.760498e-01 2.395018e-02 Class1 ## 352 Class2 6.705924e-02 9.329408e-01 Class2 ## 353 Class1 9.997610e-01 2.390348e-04 Class1 ## 354 Class1 9.366396e-02 9.063360e-01 Class2 ## 355 Class2 9.319175e-03 9.906808e-01 Class2 ## 356 Class1 9.967087e-01 3.291322e-03 Class1 ## 357 Class2 4.388100e-03 9.956119e-01 Class2 ## 358 Class1 9.888784e-01 1.112161e-02 Class1 ## 359 Class1 6.212697e-01 3.787303e-01 Class1 ## 360 Class2 3.660464e-04 9.996340e-01 Class2 ## 361 Class1 9.994841e-01 5.159239e-04 Class1 ## 362 Class1 9.589573e-01 4.104268e-02 Class1 ## 363 Class2 2.696058e-01 7.303942e-01 Class2 ## 364 Class2 6.525141e-02 9.347486e-01 Class2 ## 365 Class1 9.711997e-01 2.880030e-02 Class1 ## 366 Class1 9.999104e-01 8.963980e-05 Class1 ## 367 Class1 8.572212e-01 1.427788e-01 Class1 ## 368 Class2 1.401020e-05 9.999860e-01 Class2 ## 369 Class1 9.876044e-01 1.239563e-02 Class1 ## 370 Class2 5.391074e-01 4.608926e-01 Class1 ## 371 Class1 7.680658e-01 2.319342e-01 Class1 ## 372 Class1 9.956330e-01 4.366955e-03 Class1 ## 373 Class2 4.699016e-01 5.300984e-01 Class2 ## 374 Class1 9.273910e-01 7.260897e-02 Class1 ## 375 Class1 9.966243e-01 3.375711e-03 Class1 ## 376 Class1 9.976469e-01 2.353108e-03 Class1 ## 377 Class2 3.465621e-01 6.534379e-01 Class2 ## 378 Class2 8.551751e-04 9.991448e-01 Class2 ## 379 Class1 9.992609e-01 7.390657e-04 Class1 ## 380 Class1 8.981958e-01 1.018042e-01 Class1 ## 381 Class2 3.045435e-01 6.954565e-01 Class2 ## 382 Class1 9.967702e-01 3.229761e-03 Class1 ## 383 Class2 3.858411e-04 9.996142e-01 Class2 ## 384 Class2 9.960875e-02 9.003913e-01 Class2 ## 385 Class2 7.448033e-02 9.255197e-01 Class2 ## 386 Class1 9.967863e-01 3.213667e-03 Class1 ## 387 Class1 9.538964e-01 4.610359e-02 Class1 ## 388 Class1 2.754466e-01 7.245534e-01 Class2 ## 389 Class2 1.842538e-01 8.157462e-01 Class2 ## 390 Class1 9.957369e-01 4.263105e-03 Class1 ## 391 Class2 5.925702e-02 9.407430e-01 Class2 ## 392 Class1 9.995413e-01 4.587143e-04 Class1 ## 393 Class1 9.850931e-01 1.490691e-02 Class1 ## 394 Class2 3.054019e-04 9.996946e-01 Class2 ## 395 Class2 6.576574e-04 9.993423e-01 Class2 ## 396 Class2 3.619922e-01 6.380078e-01 Class2 ## 397 Class2 1.799232e-01 8.200768e-01 Class2 ## 398 Class1 9.976688e-01 2.331175e-03 Class1 ## 399 Class2 7.233310e-04 9.992767e-01 Class2 ## 400 Class1 9.993188e-01 6.812173e-04 Class1 ## 401 Class1 9.986897e-01 1.310336e-03 Class1 ## 402 Class1 9.855755e-01 1.442446e-02 Class1 ## 403 Class1 9.978379e-01 2.162137e-03 Class1 ## 404 Class1 8.816057e-01 1.183943e-01 Class1 ## 405 Class1 9.953082e-01 4.691808e-03 Class1 ## 406 Class1 9.470567e-01 5.294331e-02 Class1 ## 407 Class2 3.247223e-03 9.967528e-01 Class2 ## 408 Class2 1.348578e-01 8.651422e-01 Class2 ## 409 Class1 9.999209e-01 7.906366e-05 Class1 ## 410 Class2 8.533897e-01 1.466103e-01 Class1 ## 411 Class2 3.410674e-01 6.589326e-01 Class2 ## 412 Class2 8.610228e-01 1.389772e-01 Class1 ## 413 Class1 9.869813e-01 1.301868e-02 Class1 ## 414 Class2 3.607902e-01 6.392098e-01 Class2 ## 415 Class1 8.076639e-01 1.923361e-01 Class1 ## 416 Class2 1.073360e-03 9.989266e-01 Class2 ## 417 Class2 6.441835e-03 9.935582e-01 Class2 ## 418 Class1 9.238690e-01 7.613105e-02 Class1 ## 419 Class1 9.417450e-01 5.825501e-02 Class1 ## 420 Class1 9.899618e-01 1.003817e-02 Class1 ## 421 Class2 7.334798e-02 9.266520e-01 Class2 ## 422 Class2 7.371977e-02 9.262802e-01 Class2 ## 423 Class1 9.664979e-01 3.350207e-02 Class1 ## 424 Class2 5.704475e-02 9.429552e-01 Class2 ## 425 Class2 3.439244e-03 9.965608e-01 Class2 ## 426 Class2 8.484827e-02 9.151517e-01 Class2 ## 427 Class1 6.703457e-01 3.296543e-01 Class1 ## 428 Class2 6.914664e-01 3.085336e-01 Class1 ## 429 Class2 3.096499e-03 9.969035e-01 Class2 ## 430 Class1 9.945145e-01 5.485533e-03 Class1 ## 431 Class2 2.518548e-02 9.748145e-01 Class2 ## 432 Class1 9.971522e-01 2.847764e-03 Class1 ## 433 Class2 4.278915e-02 9.572109e-01 Class2 ## 434 Class1 9.896614e-01 1.033858e-02 Class1 ## 435 Class1 8.740457e-01 1.259543e-01 Class1 ## 436 Class1 9.993288e-01 6.711733e-04 Class1 ## 437 Class2 6.007991e-01 3.992009e-01 Class1 ## 438 Class1 8.819697e-01 1.180303e-01 Class1 ## 439 Class2 6.367078e-04 9.993633e-01 Class2 ## 440 Class1 9.828755e-01 1.712448e-02 Class1 ## 441 Class1 9.786333e-01 2.136674e-02 Class1 ## 442 Class2 6.797108e-01 3.202892e-01 Class1 ## 443 Class1 9.825185e-01 1.748150e-02 Class1 ## 444 Class1 4.449014e-01 5.550986e-01 Class2 ## 445 Class1 7.242637e-01 2.757363e-01 Class1 ## 446 Class1 9.968111e-01 3.188885e-03 Class1 ## 447 Class1 9.695749e-01 3.042514e-02 Class1 ## 448 Class2 3.345763e-02 9.665424e-01 Class2 ## 449 Class2 1.321441e-04 9.998679e-01 Class2 ## 450 Class1 9.928480e-01 7.151964e-03 Class1 ## 451 Class1 8.991757e-02 9.100824e-01 Class2 ## 452 Class2 1.039585e-02 9.896042e-01 Class2 ## 453 Class1 9.919739e-01 8.026119e-03 Class1 ## 454 Class1 6.176158e-01 3.823842e-01 Class1 ## 455 Class1 6.587248e-01 3.412752e-01 Class1 ## 456 Class1 2.332712e-01 7.667288e-01 Class2 ## 457 Class1 5.571984e-01 4.428016e-01 Class1 ## 458 Class1 8.159535e-01 1.840465e-01 Class1 ## 459 Class1 9.436255e-01 5.637448e-02 Class1 ## 460 Class2 1.353971e-03 9.986460e-01 Class2 ## 461 Class2 2.475278e-04 9.997525e-01 Class2 ## 462 Class1 9.876524e-01 1.234763e-02 Class1 ## 463 Class1 8.822350e-01 1.177650e-01 Class1 ## 464 Class2 6.505457e-01 3.494543e-01 Class1 ## 465 Class1 8.486386e-01 1.513614e-01 Class1 ## 466 Class2 1.303776e-01 8.696224e-01 Class2 ## 467 Class1 3.761876e-01 6.238124e-01 Class2 ## 468 Class1 9.833771e-01 1.662292e-02 Class1 ## 469 Class1 2.904065e-01 7.095935e-01 Class2 ## 470 Class1 9.998974e-01 1.026120e-04 Class1 ## 471 Class1 9.748350e-01 2.516500e-02 Class1 ## 472 Class1 9.794218e-01 2.057816e-02 Class1 ## 473 Class1 9.707829e-01 2.921708e-02 Class1 ## 474 Class2 1.802205e-01 8.197795e-01 Class2 ## 475 Class1 4.537479e-02 9.546252e-01 Class2 ## 476 Class2 2.413827e-01 7.586173e-01 Class2 ## 477 Class1 2.194477e-01 7.805523e-01 Class2 ## 478 Class2 7.363009e-03 9.926370e-01 Class2 ## 479 Class1 9.988227e-01 1.177294e-03 Class1 ## 480 Class2 7.479419e-01 2.520581e-01 Class1 ## 481 Class2 2.181312e-01 7.818688e-01 Class2 ## 482 Class1 9.484841e-01 5.151590e-02 Class1 ## 483 Class1 9.738766e-01 2.612340e-02 Class1 ## 484 Class2 1.277567e-02 9.872243e-01 Class2 ## 485 Class2 1.316514e-01 8.683486e-01 Class2 ## 486 Class2 1.307568e-03 9.986924e-01 Class2 ## 487 Class2 1.794262e-07 9.999998e-01 Class2 ## 488 Class1 9.904621e-01 9.537923e-03 Class1 ## 489 Class2 6.036590e-03 9.939634e-01 Class2 ## 490 Class2 7.528744e-01 2.471256e-01 Class1 ## 491 Class2 2.724616e-02 9.727538e-01 Class2 ## 492 Class1 9.998638e-01 1.362299e-04 Class1 ## 493 Class1 9.854221e-01 1.457794e-02 Class1 ## 494 Class1 3.752641e-01 6.247359e-01 Class2 ## 495 Class1 7.153528e-02 9.284647e-01 Class2 ## 496 Class2 6.451378e-01 3.548622e-01 Class1 ## 497 Class2 5.866325e-02 9.413367e-01 Class2 ## 498 Class2 2.014645e-01 7.985355e-01 Class2 ## 499 Class2 8.261814e-02 9.173819e-01 Class2 ## 500 Class1 9.425177e-01 5.748227e-02 Class1 ``` --- # Performance metrics ```r two_class_example %>% accuracy(truth = truth, estimate = predicted) ``` ``` ## # A tibble: 1 x 3 ## .metric .estimator .estimate ## <chr> <chr> <dbl> ## 1 accuracy binary 0.838 ``` ```r two_class_example %>% specificity(truth = truth, estimate = predicted) ``` ``` ## # A tibble: 1 x 3 ## .metric .estimator .estimate ## <chr> <chr> <dbl> ## 1 spec binary 0.793 ``` ```r two_class_example %>% sensitivity(truth = truth, estimate = predicted) ``` ``` ## # A tibble: 1 x 3 ## .metric .estimator .estimate ## <chr> <chr> <dbl> ## 1 sens binary 0.880 ``` --- # Performance metrics ```r my_metrics <- metric_set(accuracy, specificity, sensitivity) two_class_example %>% my_metrics(truth = truth, estimate = predicted) ``` ``` ## # A tibble: 3 x 3 ## .metric .estimator .estimate ## <chr> <chr> <dbl> ## 1 accuracy binary 0.838 ## 2 spec binary 0.793 ## 3 sens binary 0.880 ``` --- # Performance metrics ```r my_metrics <- metric_set(accuracy, specificity, sensitivity) two_class_example %>% group_by(group = row_number() %% 3) %>% my_metrics(truth = truth, estimate = predicted) ``` ``` ## # A tibble: 9 x 4 ## group .metric .estimator .estimate ## <dbl> <chr> <chr> <dbl> ## 1 0 accuracy binary 0.843 ## 2 1 accuracy binary 0.826 ## 3 2 accuracy binary 0.844 ## 4 0 spec binary 0.816 ## 5 1 spec binary 0.782 ## 6 2 spec binary 0.785 ## 7 0 sens binary 0.867 ## 8 1 sens binary 0.875 ## 9 2 sens binary 0.898 ``` --- # Performance metrics ```r two_class_example %>% roc_curve(truth, Class1) ``` ``` ## # A tibble: 502 x 3 ## .threshold specificity sensitivity ## <dbl> <dbl> <dbl> ## 1 -Inf 0 1 ## 2 1.79e-7 0 1 ## 3 4.50e-6 0.00413 1 ## 4 5.81e-6 0.00826 1 ## 5 5.92e-6 0.0124 1 ## 6 1.22e-5 0.0165 1 ## 7 1.40e-5 0.0207 1 ## 8 1.43e-5 0.0248 1 ## 9 2.38e-5 0.0289 1 ## 10 3.30e-5 0.0331 1 ## # … with 492 more rows ``` --- # Performance metrics ```r roc_curve(two_class_example, truth, Class1) %>% autoplot() ``` <img src="index_files/figure-html/unnamed-chunk-19-1.png" width="700px" style="display: block; margin: auto;" /> --- # Performance metrics ```r two_class_example %>% group_by(group = row_number() %% 3) %>% roc_curve(truth, Class1) %>% autoplot() ``` <img src="index_files/figure-html/unnamed-chunk-20-1.png" width="700px" style="display: block; margin: auto;" /> --- background-image: url(diagrams/full-game-rsample.png) background-position: center background-size: contain --- # rsample Spending our data carefully and efficiently --- # penguins ```r library(palmerpenguins) penguins ``` ``` ## # A tibble: 344 x 8 ## species island bill_length_mm bill_depth_mm flipper_length_mm body_mass_g ## <fct> <fct> <dbl> <dbl> <int> <int> ## 1 Adelie Torgersen 39.1 18.7 181 3750 ## 2 Adelie Torgersen 39.5 17.4 186 3800 ## 3 Adelie Torgersen 40.3 18 195 3250 ## 4 Adelie Torgersen NA NA NA NA ## 5 Adelie Torgersen 36.7 19.3 193 3450 ## 6 Adelie Torgersen 39.3 20.6 190 3650 ## 7 Adelie Torgersen 38.9 17.8 181 3625 ## 8 Adelie Torgersen 39.2 19.6 195 4675 ## 9 Adelie Torgersen 34.1 18.1 193 3475 ## 10 Adelie Torgersen 42 20.2 190 4250 ## # … with 334 more rows, and 2 more variables: sex <fct>, year <int> ``` --- # Validation split ```r set.seed(1234) penguins_split <- initial_split(penguins, prop = 0.7, strata = species) penguins ``` ``` ## # A tibble: 344 x 8 ## species island bill_length_mm bill_depth_mm flipper_length_mm body_mass_g ## <fct> <fct> <dbl> <dbl> <int> <int> ## 1 Adelie Torgersen 39.1 18.7 181 3750 ## 2 Adelie Torgersen 39.5 17.4 186 3800 ## 3 Adelie Torgersen 40.3 18 195 3250 ## 4 Adelie Torgersen NA NA NA NA ## 5 Adelie Torgersen 36.7 19.3 193 3450 ## 6 Adelie Torgersen 39.3 20.6 190 3650 ## 7 Adelie Torgersen 38.9 17.8 181 3625 ## 8 Adelie Torgersen 39.2 19.6 195 4675 ## 9 Adelie Torgersen 34.1 18.1 193 3475 ## 10 Adelie Torgersen 42 20.2 190 4250 ## # … with 334 more rows, and 2 more variables: sex <fct>, year <int> ``` --- # Validation split ```r set.seed(1234) penguins_split <- initial_split(penguins, prop = 0.7, strata = species) penguins_train <- training(penguins_split) penguins_test <- testing(penguins_split) penguins_train ``` ``` ## # A tibble: 239 x 8 ## species island bill_length_mm bill_depth_mm flipper_length_mm body_mass_g ## <fct> <fct> <dbl> <dbl> <int> <int> ## 1 Adelie Torgersen 39.5 17.4 186 3800 ## 2 Adelie Torgersen 40.3 18 195 3250 ## 3 Adelie Torgersen NA NA NA NA ## 4 Adelie Torgersen 39.3 20.6 190 3650 ## 5 Adelie Torgersen 39.2 19.6 195 4675 ## 6 Adelie Torgersen 34.1 18.1 193 3475 ## 7 Adelie Torgersen 42 20.2 190 4250 ## 8 Adelie Torgersen 37.8 17.1 186 3300 ## 9 Adelie Torgersen 41.1 17.6 182 3200 ## 10 Adelie Torgersen 38.6 21.2 191 3800 ## # … with 229 more rows, and 2 more variables: sex <fct>, year <int> ``` --- # Bootstrapping ```r penguins_boots <- bootstraps(penguins_train, times = 1000) penguins_boots ``` ``` ## # Bootstrap sampling ## # A tibble: 1,000 x 2 ## splits id ## <list> <chr> ## 1 <split [239/96]> Bootstrap0001 ## 2 <split [239/90]> Bootstrap0002 ## 3 <split [239/91]> Bootstrap0003 ## 4 <split [239/84]> Bootstrap0004 ## 5 <split [239/92]> Bootstrap0005 ## 6 <split [239/92]> Bootstrap0006 ## 7 <split [239/97]> Bootstrap0007 ## 8 <split [239/85]> Bootstrap0008 ## 9 <split [239/91]> Bootstrap0009 ## 10 <split [239/83]> Bootstrap0010 ## # … with 990 more rows ``` --- # Bootstrapping ```r penguins_boots <- bootstraps(penguins_train, times = 1000) ``` ```r lobstr::obj_size(penguins_train) ``` ``` ## 12,640 B ``` ```r lobstr::obj_size(penguins_boots) ``` ``` ## 1,774,664 B ``` --- # Cross-Validation ```r penguins_boots <- vfold_cv(penguins_train) penguins_boots ``` ``` ## # 10-fold cross-validation ## # A tibble: 10 x 2 ## splits id ## <list> <chr> ## 1 <split [215/24]> Fold01 ## 2 <split [215/24]> Fold02 ## 3 <split [215/24]> Fold03 ## 4 <split [215/24]> Fold04 ## 5 <split [215/24]> Fold05 ## 6 <split [215/24]> Fold06 ## 7 <split [215/24]> Fold07 ## 8 <split [215/24]> Fold08 ## 9 <split [215/24]> Fold09 ## 10 <split [216/23]> Fold10 ``` --- background-image: url(diagrams/full-game-recipes.png) background-position: center background-size: contain --- # recipes ## What happens to the data between `read_data()` and `fit_model()`? --- ## Prices of 54,000 round-cut diamonds ```r library(ggplot2) diamonds ``` ``` ## # A tibble: 53,940 x 10 ## carat cut color clarity depth table price x y z ## <dbl> <ord> <ord> <ord> <dbl> <dbl> <int> <dbl> <dbl> <dbl> ## 1 0.23 Ideal E SI2 61.5 55 326 3.95 3.98 2.43 ## 2 0.21 Premium E SI1 59.8 61 326 3.89 3.84 2.31 ## 3 0.23 Good E VS1 56.9 65 327 4.05 4.07 2.31 ## 4 0.29 Premium I VS2 62.4 58 334 4.2 4.23 2.63 ## 5 0.31 Good J SI2 63.3 58 335 4.34 4.35 2.75 ## 6 0.24 Very Good J VVS2 62.8 57 336 3.94 3.96 2.48 ## 7 0.24 Very Good I VVS1 62.3 57 336 3.95 3.98 2.47 ## 8 0.26 Very Good H SI1 61.9 55 337 4.07 4.11 2.53 ## 9 0.22 Fair E VS2 65.1 61 337 3.87 3.78 2.49 ## 10 0.23 Very Good H VS1 59.4 61 338 4 4.05 2.39 ## # … with 53,930 more rows ``` --- ## Formula expression in modeling ```r model <- lm(price ~ cut:color + carat + log(depth), data = diamonds) ``` - Select .orange[outcome] & .blue[predictors] --- ## Formula expression in modeling ```r model <- lm(price ~ cut:color + carat + log(depth), data = diamonds) ``` - Select outcome & predictors - .orange[Operators] to matrix of predictors --- ## Formula expression in modeling ```r model <- lm(price ~ cut:color + carat + log(depth), data = diamonds) ``` - Select outcome & predictors - Operators to matrix of predictors - .orange[Inline functions] --- ## Work under the hood - model.matrix ```r model.matrix(price ~ cut:color + carat + log(depth) + table, data = diamonds) ``` ``` ## Rows: 53,940 ## Columns: 39 ## $ `(Intercept)` <dbl> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, … ## $ carat <dbl> 0.23, 0.21, 0.23, 0.29, 0.31, 0.24, 0.24, 0.26, … ## $ `log(depth)` <dbl> 4.119037, 4.091006, 4.041295, 4.133565, 4.147885… ## $ table <dbl> 55, 61, 65, 58, 58, 57, 57, 55, 61, 61, 55, 56, … ## $ `cutFair:colorD` <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, … ## $ `cutGood:colorD` <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, … ## $ `cutVery Good:colorD` <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, … ## $ `cutPremium:colorD` <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, … ## $ `cutIdeal:colorD` <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, … ## $ `cutFair:colorE` <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, … ## $ `cutGood:colorE` <dbl> 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, … ## $ `cutVery Good:colorE` <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, … ## $ `cutPremium:colorE` <dbl> 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 1, … ## $ `cutIdeal:colorE` <dbl> 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, … ## $ `cutFair:colorF` <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, … ## $ `cutGood:colorF` <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, … ## $ `cutVery Good:colorF` <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, … ## $ `cutPremium:colorF` <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, … ## $ `cutIdeal:colorF` <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, … ## $ `cutFair:colorG` <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, … ## $ `cutGood:colorG` <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, … ## $ `cutVery Good:colorG` <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, … ## $ `cutPremium:colorG` <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, … ## $ `cutIdeal:colorG` <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, … ## $ `cutFair:colorH` <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, … ## $ `cutGood:colorH` <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, … ## $ `cutVery Good:colorH` <dbl> 0, 0, 0, 0, 0, 0, 0, 1, 0, 1, 0, 0, 0, 0, 0, 0, … ## $ `cutPremium:colorH` <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, … ## $ `cutIdeal:colorH` <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, … ## $ `cutFair:colorI` <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, … ## $ `cutGood:colorI` <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, … ## $ `cutVery Good:colorI` <dbl> 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, … ## $ `cutPremium:colorI` <dbl> 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, … ## $ `cutIdeal:colorI` <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, … ## $ `cutFair:colorJ` <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, … ## $ `cutGood:colorJ` <dbl> 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, … ## $ `cutVery Good:colorJ` <dbl> 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, … ## $ `cutPremium:colorJ` <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, … ## $ `cutIdeal:colorJ` <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 1, 0, 0, … ``` --- ## Downsides - **Tedious typing with many variables** --- ## Downsides - Tedious typing with many variables - **Functions have to manually be applied to each variable** ```r lm(y ~ log(x01) + log(x02) + log(x03) + log(x04) + log(x05) + log(x06) + log(x07) + log(x08) + log(x09) + log(x10) + log(x11) + log(x12) + log(x13) + log(x14) + log(x15) + log(x16) + log(x17) + log(x18) + log(x19) + log(x20) + log(x21) + log(x22) + log(x23) + log(x24) + log(x25) + log(x26) + log(x27) + log(x28) + log(x29) + log(x30) + log(x31) + log(x32) + log(x33) + log(x34) + log(x35), data = dat) ``` --- ## Downsides - Tedious typing with many variables - Functions have to manually be applied to each variable - **Operations are constrained to single columns** ```r # Not possible lm(y ~ pca(x01, x02, x03, x04, x05), data = dat) ``` --- ## Downsides - Tedious typing with many variables - Functions have to manually be applied to each variable - Operations are constrained to single columns - **Everything happens at once** You can't apply multiple transformations to the same variable. --- ## Downsides - Tedious typing with many variables - Functions have to manually be applied to each variable - Operations are constrained to single columns - Everything happens at once - **Connected to the model, calculations are not saved between models** One could manually use `model.matrix` and pass the result to the modeling function. --- # Recipes New package to deal with this problem ### Benefits: - **Modular** --- # Recipes New package to deal with this problem ### Benefits: - Modular - **pipeable** --- # Recipes New package to deal with this problem ### Benefits: - Modular - pipeable - **Deferred evaluation** --- # Recipes New package to deal with this problem ### Benefits: - Modular - pipeable - Deferred evaluation - **Isolates test data from training data** --- # Recipes New package to deal with this problem ### Benefits: - Modular - pipeable - Deferred evaluation - Isolates test data from training data - **Can do things formulas can't** --- # Modularity and pipeability ```r price ~ cut + color + carat + log(depth) + table ``` Taking the formula from before we can rewrite it as the following recipe ```r rec <- recipe(price ~ cut + color + carat + depth + table, data = diamonds) %>% step_log(depth) %>% step_dummy(cut, color) ``` --- # Modularity and pipeability ```r price ~ cut + color + carat + log(depth) + table ``` Taking the formula from before we can rewrite it as the following recipe ```r rec <- recipe(price ~ cut + color + carat + depth + table, data = diamonds) %>% step_log(depth) %>% step_dummy(cut, color) ``` .orange[formula] expression to specify variables --- # Modularity and pipeability ```r price ~ cut + color + carat + log(depth) + table ``` Taking the formula from before we can rewrite it as the following recipe ```r rec <- recipe(price ~ cut + color + carat + depth + table, data = diamonds) %>% step_log(depth) %>% step_dummy(cut, color) ``` then apply .orange[log] transformation on `depth` --- # Modularity and pipeability ```r price ~ cut + color + carat + log(depth) + table ``` Taking the formula from before we can rewrite it as the following recipe ```r rec <- recipe(price ~ cut + color + carat + depth + table, data = diamonds) %>% step_log(depth) %>% step_dummy(cut, color) ``` lastly we create .orange[dummy variables] from `cut` and `color` --- ## Deferred evaluation If we look at the recipe we created we don't see a dataset, but instead, we see a specification ```r rec ``` ``` ## Data Recipe ## ## Inputs: ## ## role #variables ## outcome 1 ## predictor 5 ## ## Operations: ## ## Log transformation on depth ## Dummy variables from cut, color ``` --- ## Deferred evaluation **recipes** gives a specification of the intent of what we want to do. No calculations have been carried out yet. First, we need to `prep()` the recipe. This will calculate the sufficient statistics needed to perform each of the steps. ```r prepped_rec <- prep(rec) ``` --- ## Deferred evaluation ```r prepped_rec ``` ``` ## Data Recipe ## ## Inputs: ## ## role #variables ## outcome 1 ## predictor 5 ## ## Training data contained 53940 data points and no missing data. ## ## Operations: ## ## Log transformation on depth [trained] ## Dummy variables from cut, color [trained] ``` --- # Baking After we have prepped the recipe we can `bake()` it to apply all the transformations ```r bake(prepped_rec, new_data = diamonds) ``` ``` ## Rows: 53,940 ## Columns: 14 ## $ carat <dbl> 0.23, 0.21, 0.23, 0.29, 0.31, 0.24, 0.24, 0.26, 0.22, 0.23, 0.… ## $ depth <dbl> 4.119037, 4.091006, 4.041295, 4.133565, 4.147885, 4.139955, 4.… ## $ table <dbl> 55, 61, 65, 58, 58, 57, 57, 55, 61, 61, 55, 56, 61, 54, 62, 58… ## $ price <int> 326, 326, 327, 334, 335, 336, 336, 337, 337, 338, 339, 340, 34… ## $ cut_1 <dbl> 0.6324555, 0.3162278, -0.3162278, 0.3162278, -0.3162278, 0.000… ## $ cut_2 <dbl> 0.5345225, -0.2672612, -0.2672612, -0.2672612, -0.2672612, -0.… ## $ cut_3 <dbl> 3.162278e-01, -6.324555e-01, 6.324555e-01, -6.324555e-01, 6.32… ## $ cut_4 <dbl> 0.1195229, -0.4780914, -0.4780914, -0.4780914, -0.4780914, 0.7… ## $ color_1 <dbl> -3.779645e-01, -3.779645e-01, -3.779645e-01, 3.779645e-01, 5.6… ## $ color_2 <dbl> 9.690821e-17, 9.690821e-17, 9.690821e-17, 0.000000e+00, 5.4554… ## $ color_3 <dbl> 4.082483e-01, 4.082483e-01, 4.082483e-01, -4.082483e-01, 4.082… ## $ color_4 <dbl> -0.5640761, -0.5640761, -0.5640761, -0.5640761, 0.2417469, 0.2… ## $ color_5 <dbl> 4.364358e-01, 4.364358e-01, 4.364358e-01, -4.364358e-01, 1.091… ## $ color_6 <dbl> -0.19738551, -0.19738551, -0.19738551, -0.19738551, 0.03289758… ``` --- # Baking / Juicing Since the dataset is already calculated after running `prep()` can we use `juice()` to extract it ```r juice(prepped_rec) ``` ``` ## Rows: 53,940 ## Columns: 14 ## $ carat <dbl> 0.23, 0.21, 0.23, 0.29, 0.31, 0.24, 0.24, 0.26, 0.22, 0.23, 0.… ## $ depth <dbl> 4.119037, 4.091006, 4.041295, 4.133565, 4.147885, 4.139955, 4.… ## $ table <dbl> 55, 61, 65, 58, 58, 57, 57, 55, 61, 61, 55, 56, 61, 54, 62, 58… ## $ price <int> 326, 326, 327, 334, 335, 336, 336, 337, 337, 338, 339, 340, 34… ## $ cut_1 <dbl> 0.6324555, 0.3162278, -0.3162278, 0.3162278, -0.3162278, 0.000… ## $ cut_2 <dbl> 0.5345225, -0.2672612, -0.2672612, -0.2672612, -0.2672612, -0.… ## $ cut_3 <dbl> 3.162278e-01, -6.324555e-01, 6.324555e-01, -6.324555e-01, 6.32… ## $ cut_4 <dbl> 0.1195229, -0.4780914, -0.4780914, -0.4780914, -0.4780914, 0.7… ## $ color_1 <dbl> -3.779645e-01, -3.779645e-01, -3.779645e-01, 3.779645e-01, 5.6… ## $ color_2 <dbl> 9.690821e-17, 9.690821e-17, 9.690821e-17, 0.000000e+00, 5.4554… ## $ color_3 <dbl> 4.082483e-01, 4.082483e-01, 4.082483e-01, -4.082483e-01, 4.082… ## $ color_4 <dbl> -0.5640761, -0.5640761, -0.5640761, -0.5640761, 0.2417469, 0.2… ## $ color_5 <dbl> 4.364358e-01, 4.364358e-01, 4.364358e-01, -4.364358e-01, 1.091… ## $ color_6 <dbl> -0.19738551, -0.19738551, -0.19738551, -0.19738551, 0.03289758… ``` --- .center[ # recipes workflow ] <br> <br> <br> .huge[ .center[ ```r recipe -> prepare -> bake/juice (define) -> (estimate) -> (apply) ``` ] ] --- ## Isolates test & training data When working with data for predictive modeling it is important to make sure any information from the test data leaks into the training data. This is avoided by using **recipes** by making sure you only prep the recipe with the training dataset. --- # Can do things formulas can't --- # selectors .pull-left[ It can be annoying to manually specify variables by name. The use of selectors can greatly help you! ] .pull-right[ ```r rec <- recipe(price ~ ., data = diamonds) %>% step_dummy(all_nominal()) %>% step_zv(all_numeric()) %>% step_center(all_predictors()) ``` ] --- # selectors .pull-left[ .orange[`all_nominal()`] is used to select all the nominal variables. ] .pull-right[ ```r rec <- recipe(price ~ ., data = diamonds) %>% step_dummy(all_nominal()) %>% step_zv(all_numeric()) %>% step_center(all_predictors()) ``` ] --- # selectors .pull-left[ .orange[`all_numeric()`] is used to select all the numeric variables. Even the ones generated by .blue[`step_dummy()`] ] .pull-right[ ```r rec <- recipe(price ~ ., data = diamonds) %>% step_dummy(all_nominal()) %>% step_zv(all_numeric()) %>% step_center(all_predictors()) ``` ] --- # selectors .pull-left[ .orange[`all_predictors()`] is used to select all predictor variables. Will not break even if a variable is removed with .blue[`step_zv()`] ] .pull-right[ ```r rec <- recipe(price ~ ., data = diamonds) %>% step_dummy(all_nominal()) %>% step_zv(all_numeric()) %>% step_center(all_predictors()) ``` ] --- # Roles .pull-left[ .orange[`update_role()`] can be used to give variables roles. That then can be selected with .blue[`has_role()`] Roles can also be set with `role = ` argument inside steps ] .pull-right[ ```r rec <- recipe(price ~ ., data = diamonds) %>% update_role(x, y, z, new_role = "size") %>% step_log(has_role("size")) %>% step_dummy(all_nominal()) %>% step_zv(all_numeric()) %>% step_center(all_predictors()) ``` ] --- ## PCA extraction ```r rec <- recipe(price ~ ., data = diamonds) %>% step_dummy(all_nominal()) %>% step_scale(all_predictors()) %>% step_center(all_predictors()) %>% step_pca(all_predictors(), threshold = 0.8) ``` You can also write a recipe that extract enough .orange[principal components] to explain .blue[80% of the variance] Loadings will be kept in the prepped recipe to make sure other datasets are transformed correctly --- background-image: url(diagrams/full-game-workflows.png) background-position: center background-size: contain --- # Workflows Simple package that helps us formulate more about what happens to our model. Main functions are `workflow()`, `add_model()`, `add_formula()` or `add_variables()` (we will see `add_recipe()` later in the course) ```r library(workflows) linear_wf <- workflow() %>% add_model(linear_spec) %>% add_formula(mpg ~ disp + hp + wt) ``` --- # Workflows This allows up to combine the model with what variables it should expect .pull-left[ ```r library(workflows) linear_wf <- workflow() %>% add_model(linear_spec) %>% add_formula(mpg ~ disp + hp + wt) linear_wf ``` ] .pull-right[ ``` ## ══ Workflow ════════════════════════════════════════════════════════════════════ ## Preprocessor: Formula ## Model: linear_reg() ## ## ── Preprocessor ──────────────────────────────────────────────────────────────── ## mpg ~ disp + hp + wt ## ## ── Model ─────────────────────────────────────────────────────────────────────── ## Linear Regression Model Specification (regression) ## ## Computational engine: lm ``` ] --- `add_variables()` allows for a different way of specifying the the response and predictors in our model # Workflows .pull-left[ ```r library(workflows) linear_wf <- workflow() %>% add_model(linear_spec) %>% add_variables(outcomes = mpg, predictors = c(disp, hp, wt)) linear_wf ``` ] .pull-right[ ``` ## ══ Workflow ════════════════════════════════════════════════════════════════════ ## Preprocessor: Variables ## Model: linear_reg() ## ## ── Preprocessor ──────────────────────────────────────────────────────────────── ## Outcomes: mpg ## Predictors: c(disp, hp, wt) ## ## ── Model ─────────────────────────────────────────────────────────────────────── ## Linear Regression Model Specification (regression) ## ## Computational engine: lm ``` ] --- # Workflows You can use a `workflow` just like a parsnip object and fit it directly ```r fit(linear_wf, data = mtcars) ``` ``` ## ══ Workflow [trained] ══════════════════════════════════════════════════════════ ## Preprocessor: Variables ## Model: linear_reg() ## ## ── Preprocessor ──────────────────────────────────────────────────────────────── ## Outcomes: mpg ## Predictors: c(disp, hp, wt) ## ## ── Model ─────────────────────────────────────────────────────────────────────── ## ## Call: ## stats::lm(formula = ..y ~ ., data = data) ## ## Coefficients: ## (Intercept) disp hp wt ## 37.105505 -0.000937 -0.031157 -3.800891 ``` --- background-image: url(diagrams/full-game-tune.png) background-position: center background-size: contain --- # Tune We introduce the **tune** package. This package helps us fit many models in a controlled manner in the tidymodels framework. It relies heavily on parsnip and rsample --- # Tune We can use `fit_resamples()` to fit the workflow we created within each resample ```r library(tune) mtcars_folds <- vfold_cv(mtcars, v = 4) linear_fold_fits <- fit_resamples( linear_wf, resamples = mtcars_folds ) ``` --- # Tune The results of this resampling comes as a data.frame ```r linear_fold_fits ``` ``` ## # Resampling results ## # 4-fold cross-validation ## # A tibble: 4 x 4 ## splits id .metrics .notes ## <list> <chr> <list> <list> ## 1 <split [24/8]> Fold1 <tibble [2 × 4]> <tibble [0 × 1]> ## 2 <split [24/8]> Fold2 <tibble [2 × 4]> <tibble [0 × 1]> ## 3 <split [24/8]> Fold3 <tibble [2 × 4]> <tibble [0 × 1]> ## 4 <split [24/8]> Fold4 <tibble [2 × 4]> <tibble [0 × 1]> ``` --- # Tune `collect_metrics()` can be used to extract the CV estimate ```r collect_metrics(linear_fold_fits) ``` ``` ## # A tibble: 2 x 6 ## .metric .estimator mean n std_err .config ## <chr> <chr> <dbl> <int> <dbl> <chr> ## 1 rmse standard 2.66 4 0.500 Preprocessor1_Model1 ## 2 rsq standard 0.832 4 0.0514 Preprocessor1_Model1 ``` --- # Tune Setting `summarize = FALSE` in `collect_metrics()` Allows us the see the individual performance metrics for each fold ```r collect_metrics(linear_fold_fits, summarize = FALSE) ``` ``` ## # A tibble: 8 x 5 ## id .metric .estimator .estimate .config ## <chr> <chr> <chr> <dbl> <chr> ## 1 Fold1 rmse standard 2.76 Preprocessor1_Model1 ## 2 Fold1 rsq standard 0.857 Preprocessor1_Model1 ## 3 Fold2 rmse standard 3.92 Preprocessor1_Model1 ## 4 Fold2 rsq standard 0.733 Preprocessor1_Model1 ## 5 Fold3 rmse standard 2.49 Preprocessor1_Model1 ## 6 Fold3 rsq standard 0.772 Preprocessor1_Model1 ## 7 Fold4 rmse standard 1.49 Preprocessor1_Model1 ## 8 Fold4 rsq standard 0.965 Preprocessor1_Model1 ``` --- .pull-left[ # Tune There are some settings we can set with `control_resamples()`. One of the most handy ones (IMO) is `verbose = TRUE` ```r library(tune) linear_fold_fits <- fit_resamples( linear_wf, resamples = mtcars_folds, control = control_resamples(verbose = TRUE) ) ``` ] .pull-right[ .center[  ] ] --- # Tune We can also directly specify the metrics that are calculated within each resample ```r library(tune) linear_fold_fits <- fit_resamples( linear_wf, resamples = mtcars_folds, metrics = metric_set(rmse, rsq, mase) ) collect_metrics(linear_fold_fits) ``` ``` ## # A tibble: 3 x 6 ## .metric .estimator mean n std_err .config ## <chr> <chr> <dbl> <int> <dbl> <chr> ## 1 mase standard 0.441 4 0.178 Preprocessor1_Model1 ## 2 rmse standard 2.66 4 0.500 Preprocessor1_Model1 ## 3 rsq standard 0.832 4 0.0514 Preprocessor1_Model1 ``` --- background-image: url(diagrams/full-game-dials.png) background-position: center background-size: contain --- # dials What if we want to tune hyperparameters? --- # Lasso spec ```r lasso_spec <- linear_reg(mixture = 1, penalty = tune()) %>% set_mode("regression") %>% set_engine("glmnet") ``` ```r rec_spec <- recipe(mpg ~ ., data = mtcars) %>% step_normalize(all_predictors()) ``` And we combine these two into a `workflow` ```r lasso_wf <- workflow() %>% add_model(lasso_spec) %>% add_recipe(rec_spec) ``` --- # Hyperparameter Tuning We also need to specify what values of the hyperparameters we are trying to tune we want to calculate. Since the lasso model can calculate all paths at once let us get back 50 evenly spaced values of `\(\lambda\)` ```r lambda_grid <- grid_regular(penalty(), levels = 50) lambda_grid ``` ``` ## # A tibble: 50 x 1 ## penalty ## <dbl> ## 1 1 e-10 ## 2 1.60e-10 ## 3 2.56e-10 ## 4 4.09e-10 ## 5 6.55e-10 ## 6 1.05e- 9 ## 7 1.68e- 9 ## 8 2.68e- 9 ## 9 4.29e- 9 ## 10 6.87e- 9 ## # … with 40 more rows ``` --- # Hyperparameter Tuning We combine these things in `tune_grid()` which works much like `fit_resamples()` but takes a `grid` argument as well ```r tune_rs <- tune_grid( object = lasso_wf, resamples = mtcars_folds, grid = lambda_grid ) ``` --- # Hyperparameter Tuning We can see how each of the values of `\(\lambda\)` is doing with `collect_metrics()` ```r collect_metrics(tune_rs) ``` ``` ## # A tibble: 100 x 7 ## penalty .metric .estimator mean n std_err .config ## <dbl> <chr> <chr> <dbl> <int> <dbl> <chr> ## 1 1 e-10 rmse standard 3.77 4 0.702 Preprocessor1_Model01 ## 2 1 e-10 rsq standard 0.606 4 0.191 Preprocessor1_Model01 ## 3 1.60e-10 rmse standard 3.77 4 0.702 Preprocessor1_Model02 ## 4 1.60e-10 rsq standard 0.606 4 0.191 Preprocessor1_Model02 ## 5 2.56e-10 rmse standard 3.77 4 0.702 Preprocessor1_Model03 ## 6 2.56e-10 rsq standard 0.606 4 0.191 Preprocessor1_Model03 ## 7 4.09e-10 rmse standard 3.77 4 0.702 Preprocessor1_Model04 ## 8 4.09e-10 rsq standard 0.606 4 0.191 Preprocessor1_Model04 ## 9 6.55e-10 rmse standard 3.77 4 0.702 Preprocessor1_Model05 ## 10 6.55e-10 rsq standard 0.606 4 0.191 Preprocessor1_Model05 ## # … with 90 more rows ``` --- # Hyperparameter Tuning .pull-left[ And there is even a plotting method that can show us how the different values of the hyperparameter are doing ] .pull-right[ ```r autoplot(tune_rs) ``` <img src="index_files/figure-html/unnamed-chunk-77-1.png" width="700px" style="display: block; margin: auto;" /> ] --- # Hyperparameter Tuning Look at the best performing one with `show_best()` and select the best with `select_best()` ```r tune_rs %>% show_best("rmse") ``` ``` ## # A tibble: 5 x 7 ## penalty .metric .estimator mean n std_err .config ## <dbl> <chr> <chr> <dbl> <int> <dbl> <chr> ## 1 0.244 rmse standard 2.73 4 0.309 Preprocessor1_Model47 ## 2 0.391 rmse standard 2.73 4 0.285 Preprocessor1_Model48 ## 3 0.153 rmse standard 2.78 4 0.323 Preprocessor1_Model46 ## 4 0.625 rmse standard 2.79 4 0.252 Preprocessor1_Model49 ## 5 0.0954 rmse standard 2.85 4 0.313 Preprocessor1_Model45 ``` ```r best_rmse <- tune_rs %>% select_best("rmse") ``` --- # Hyperparameter Tuning Remember how the specification has `penalty = tune()`? ```r lasso_wf ``` ``` ## ══ Workflow ════════════════════════════════════════════════════════════════════ ## Preprocessor: Recipe ## Model: linear_reg() ## ## ── Preprocessor ──────────────────────────────────────────────────────────────── ## 1 Recipe Step ## ## • step_normalize() ## ## ── Model ─────────────────────────────────────────────────────────────────────── ## Linear Regression Model Specification (regression) ## ## Main Arguments: ## penalty = tune() ## mixture = 1 ## ## Computational engine: glmnet ``` --- # Hyperparameter Tuning We can update it with `finalize_workflow()` ```r final_lasso <- finalize_workflow(lasso_wf, best_rmse) final_lasso ``` ``` ## ══ Workflow ════════════════════════════════════════════════════════════════════ ## Preprocessor: Recipe ## Model: linear_reg() ## ## ── Preprocessor ──────────────────────────────────────────────────────────────── ## 1 Recipe Step ## ## • step_normalize() ## ## ── Model ─────────────────────────────────────────────────────────────────────── ## Linear Regression Model Specification (regression) ## ## Main Arguments: ## penalty = 0.244205309454865 ## mixture = 1 ## ## Computational engine: glmnet ``` --- # Hyperparameter Tuning And this finalized specification can now we can fit using the whole training data set. ```r fitted_lasso <- fit(final_lasso, mtcars) ``` --- class: center # What now? --- # Each of these packages are created to allow for easy extensions --- background-image: url(images/parsnip-extensions.png) background-position: center background-size: contain # parsnip extensions https://www.tidymodels.org/learn/develop/models/ - *discrim* discriminant analysis models - *poissonreg* Poisson regression models - *rules* rule-based models - *baguette* bagging ensemble models - *plsmod* linear projection model - *modeltime* time series forecast models - *treesnip* tree, lightGBM, and Catboost - *censored* censored regression and survival analysis models --- # broom extensions https://www.tidymodels.org/learn/develop/broom/ - *broomstick* decision tree methods - *tidytext* corpus, LDA, topic models - *sweep* time series forecasting - *broom.mixed* mixed models --- # yardstick extensions https://www.tidymodels.org/learn/develop/metrics/ --- # rsample extensions - *spatialsample* spatial resampling .center[  ] --- # recipes extensions https://www.tidymodels.org/learn/develop/recipes/ - *embed* categorical predictor embeddings - *timetk* time series data - *textrecipes* preprocessing text - *themis* unbalanced data --- # workflows extensions - *workflowsets* create a workflow set that holds multiple workflow object --- # tune extensions - *finetune* - Efficient grid search via racing with ANOVA models - Efficient grid search via racing with win/loss statistics - Optimization of model parameters via simulated annealing --- # Other - *stacks* model stacking - *probably* Post-Processing Class Probability Estimates - *butcher* reduce the size of model objects - *modeldb* Fit models inside the database - *tidypredict* predictions inside databases --- # Resources https://www.tidymodels.org/ https://www.tmwr.org/ https://juliasilge.com/ https://emilhvitfeldt.github.io/ISLR-tidymodels-labs/index.html --- class: center, middle # Thank you! ### <i class="fab fa-github "></i> [EmilHvitfeldt](https://github.com/EmilHvitfeldt/) ### <i class="fab fa-twitter "></i> [@Emil_Hvitfeldt](https://twitter.com/Emil_Hvitfeldt) ### <i class="fab fa-linkedin "></i> [emilhvitfeldt](linkedin.com/in/emilhvitfeldt/) ### <i class="fas fa-laptop "></i> [www.hvitfeldt.me](www.hvitfeldt.me) Slides created via the R package [xaringan](https://github.com/yihui/xaringan).